REST API
Overview
The majority of actions users take in the Signals GUI actions are available in our public API. The core of most integrations is the extensive library of these APIs documented in Swagger.
API Documentation
APIs are accessed from a Swagger UI site specific to your implementation of Signals. Developers have direct visibility and access to all APIs available to them through this site. The site is dynamic, meaning that new APIs are real-time documented in the Swagger UI when made available to developers. All APIs are versioned, allowing clients to connect with a specific version to mitigate the risk of breaking existing integrations.
Adherence to Standards
APIs follow the JSON:API standard which includes document structure, header content, response codes, and more. Additionally, where applicable, specific APIs follow additional standards, such as the SCIM 2.0 standard for Group and User Management APIs (System for Cross-domain Identity Management).
Authentication
Both API key and OAuth token-based authentication are supported for secure access to APIs.
API Keys
API keys are used when making authenticated requests by including the API key in the x-api-key header in the HTTP headers of the request. API keys are generated by system admins for specific users. A given tenant may have 10 users with assigned API keys.
Generating API Keys
Signals administrators have the ability to create, view, and delete API keys. In order for an API key to access the Signals API it needs be tied to an existing user in the tenant. The API access rights are determined by those of the user associated with the API key.
To add an API Key:
1. From System Configuration home page select "System Settings".
2. Click "API Key" in the left menu.
3. Select the email address to generate an API key for its the corresponding user.
4. Click Generate API Key button. A key gets generated in the API Key field as shown in the example below:
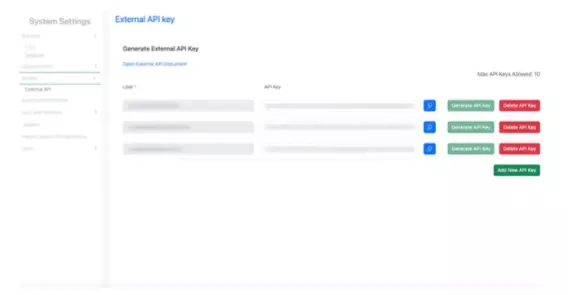
To delete an API Key:
Click on Delete API Key button, a confirmation message appears, click Delete.
OAuth Token-Based Authentication
Signals Notebook supports implicit bearer token exchange - an authentication mechanism by which a user of "an application" obtains a token from Signals Notebook after proper authentication and authorization. The end-user will be prompted to login to Signals and upon successful login will generate an authenticated bearer token. This token can then be used by "your application" to invoke Signals Notebook external APIs as the user for which the token was generated. The API access rights are determined by those of the user associated with the bearer token.
NOTE: Signals Notebook currently supports only the "implicit" grant type. https://tools.ietf.org/html/rfc6749#section-4.2
Setting up Bearer Token Exchange
Client ID Request:
To request creation of a client id contact your Customer Success Manager, your Account Manager or contact Signals support, providing the following information:
- Name of "your application" that will be accessing Signals Notebook using the bearer token. This can be any reasonably short string.
- Redirect URI(s). This is the URI(s) that the user will be redirected to once a bearer token is made available. In order to keep tight security, the redirect URI(s) must match exactly. As you develop your application integration(s), the redirect URI(s) can be updated.
- Please refer to https://tools.ietf.org/html/rfc6749#section-4.2 for more details.
Post Client ID request Steps:
1. Once a client id is made available to you, "your application" initiates a request for a token by redirecting the user to:
- <your-domain-url>/auth/oauth/authorize
- Please refer to https://tools.ietf.org/html/rfc6749#section-4.2.1 for more details.
- Signals Notebook engineering will provide values for response_type , client_id , and scope query params.
2. The user will be prompted for credentials, using their own IdP if one is set up.
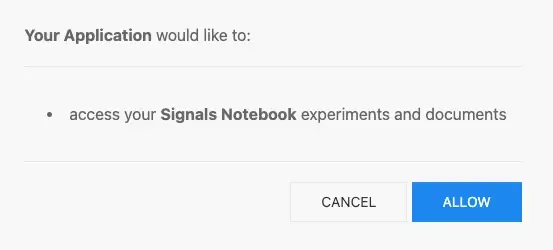
3. After successful authentication, the user will be asked to authorize "your application" to access Signals Notebook.
4. Once the user is authenticated and the user has authorized "your application", a token will be made available via the redirect URI provided in the original request (step 2). For example:
- <redirect_uri>#access_token= f60fxxxxd134d26a685737f3e6ceabe Please refer to =
- https://tools.ietf.org/html/rfc6749#section-4.2.2 for more details
5. At this point, "your application" is now responsible for extracting and caching the given access token, which can be used for accessing the external API as a bearer token in the Authorization HTTP header. For example:
- Authorization: Bearer f60fxxxxd134d26a685737f3e6ceabe
NOTE: The bearer token is valid as long as it's used in the last 30 days. After 30 days of inactivity, the token is invalidated and discarded, and the user must request for a new bearer token.
The screen shot below shows an example of the end user authentication screen provided to an external application using the application name provided during Client ID request using a bearer token exchange to authenticate against the Signals Notebook API:
Which Authentication to use?
When making use of the API for server to server integrations where there is no end-user action taking place we recommend use of the API Key. Typically you will make use of a API/System User account for these integrations as they are actions taken by the system and not an end-user. Examples of these types of integrations are automated archival of closed experiments, compliance dashboards, and external data syncs.
For integrations that involve an end-user interaction, especially those which leave an imprint in the application, such as entity modification or any action that results in an an impression in the audit trail we recommend use of the implicit bearer token exchange. This makes sure the end-users actions are accounted for and limited to their specific user permissions when interacting with the API. Examples of these types of integrations are external sample registration and data retrieval from external sources.
Limits
Rate Limits
Rate limits are set on the tenant level and limited to 1000 requests between API Tokens + Bearer Tokens per minute.
NOTE: The rate limits are subject to change to ensure consistent and responsive APIs.
Understanding API Responses
At the heart of Signals integration lies the powerful JSON:API specification. JSON:API is a widely adopted standard for designing web APIs that use JSON (JavaScript Object Notation) as the data format for communication. Understanding JSON:API is crucial for seamless integration with Signals Notebook, as it provides a structured and consistent approach to data retrieval, manipulation, and resource relationships.
Benefits of JSON:API for Integrations:
- Consistent Data Structure: JSON:API defines clear rules for structuring API responses and requests, making it easier for developers to understand and implement integrations with consistent response structures.
- Reduced Overhead: By utilizing features like resource relationships and sparse fieldsets, JSON:API minimizes data overhead, leading to decreased bandwidth usage, faster API responses, and improved overall performance.
- Efficient Resource Relationships: JSON:API excels in handling complex relationships between resources, allowing you to fetch related data in a single API call, streamlining data retrieval and eliminating the need for multiple requests.
- Inclusion of Related Resources: JSON:API enables clients to request related resources along with the main resource, reducing the need for additional API calls and optimizing performance.
- Access to Client and Server Libraries: The JSON:API is widely adopted and has many pre-existing libraries available for use to handle a lot of the heavy lifting when both creating and interpreting JSON:API requests and responses. Check out existing implementations here: https://jsonapi.org/implementations/
JSON:API Response Breakdown
When integrating with Signals Notebook, understanding the JSON:API specification and response format is essential. Here is an overview of what a JSON:API response contains:
Top-level "data" object: Represents the primary resource(s) being returned. If it's an object, it's a single resource; if it's an array, it's a collection of resources. Look for the type and id to identify the resource and check its attributes for specific data.
Relationships: Within the primary resource object, the relationships object defines links to related resources. Each relationship may have links and data that point to associated resources.
"Included" array: Contains related resources that are included in the response but are not the primary data. These resources can be referenced by the primary resource's relationships. Each included resource follows the same structure as the primary resource. To find the object in the included array you are interested in you match the type and id of the object referenced in the main data object.
"Links": Throughout the response, there may be links that provide URLs for related actions or additional information about the API. In the case of paginated API responses links to the next and previous page are also included.
Fetching an Experiment
To better understand how an entity is represented we will look at the top-level data object in the response from the GET /entities/{eid} API endpoint when used to fetch an Experiment.
API Request URL:
GET https://snb.example.com/api/rest/v1.0/entities/experiment:966a7304-4436-4f84-b56b-053c2ba2e439Response:
{
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/experiment:966a7304-4436-4f84-b56b-053c2ba2e439"
},
"data": {
"type": "entity",
"id": "experiment:966a7304-4436-4f84-b56b-053c2ba2e439",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/experiment:966a7304-4436-4f84-b56b-053c2ba2e439"
},
"attributes": {
"id": "experiment:966a7304-4436-4f84-b56b-053c2ba2e439",
"eid": "experiment:966a7304-4436-4f84-b56b-053c2ba2e439",
"name": "My First Signals Experiment",
"description": "A brand new experiment in Signals Notebook",
"createdAt": "2024-01-22T21:17:12.334Z",
"editedAt": "2024-01-22T21:27:38.958Z",
"type": "experiment",
"state": "open",
"digest": "72378008",
"fields": {
"Description": {
"value": "A brand new experiment in Signals Notebook"
},
"Name": {
"value": "My First Signals Experiment"
}
},
"flags": {
"canEdit": true
}
},
"relationships": {
"createdBy": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/users/100"
},
"data": {
"type": "user",
"id": "100"
}
},
"editedBy": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/users/100"
},
"data": {
"type": "user",
"id": "100"
}
},
"children": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/experiment:966a7304-4436-4f84-b56b-053c2ba2e439/children"
},
"data": [
{
"type": "entity",
"id": "text:18849188-9d5a-4064-a287-e4fe402028f8",
"meta": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:18849188-9d5a-4064-a287-e4fe402028f8"
}
}
},
{
"type": "entity",
"id": "text:983b313a-6a90-43cd-88ce-83b6c19f94a2",
"meta": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:983b313a-6a90-43cd-88ce-83b6c19f94a2"
}
}
},
{
"type": "entity",
"id": "text:df47fec8-d7fa-49a4-b2cf-c1468f1ad4e0",
"meta": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:df47fec8-d7fa-49a4-b2cf-c1468f1ad4e0"
}
}
},
{
"type": "entity",
"id": "chemicalDrawing:865d61d8-16c0-41c5-adb1-9d2de7999a43",
"meta": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/chemicalDrawing:865d61d8-16c0-41c5-adb1-9d2de7999a43"
}
}
},
{
"type": "entity",
"id": "samplesContainer:ca05dfb4-d4cd-4910-a486-3ebaaf7982d7",
"meta": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/samplesContainer:ca05dfb4-d4cd-4910-a486-3ebaaf7982d7"
}
}
}
]
},
"owner": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/users/100"
},
"data": {
"type": "user",
"id": "100"
}
},
"pdf": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/experiment:966a7304-4436-4f84-b56b-053c2ba2e439/pdf"
}
}
}
},
"included": [
{
"type": "user",
"id": "100",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/users/100"
},
"attributes": {
"userId": "100",
"userName": "example.user@revvity.com",
"flags": {
"isSystemStandardUser": true
},
"email": "example.user@revvity.com",
"firstName": "Example",
"lastName": "User",
"isEnabled": true
},
"relationships": {
"systemGroups": {
"links": {
"self": "https://snb.example.com/api/rest/v1.0/users/100/systemGroups"
}
}
}
},
{
"type": "entity",
"id": "text:983b313a-6a90-43cd-88ce-83b6c19f94a2",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:983b313a-6a90-43cd-88ce-83b6c19f94a2"
},
"attributes": {
"type": "text",
"eid": "text:983b313a-6a90-43cd-88ce-83b6c19f94a2",
"name": "Experiment Description",
"digest": "12062137",
"fields": {
"Description": {
"value": ""
},
"Name": {
"value": "Experiment Description"
}
}
}
},
{
"type": "entity",
"id": "chemicalDrawing:865d61d8-16c0-41c5-adb1-9d2de7999a43",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/chemicalDrawing:865d61d8-16c0-41c5-adb1-9d2de7999a43"
},
"attributes": {
"type": "chemicalDrawing",
"eid": "chemicalDrawing:865d61d8-16c0-41c5-adb1-9d2de7999a43",
"name": "H20 Chemical Drawing",
"digest": "51219370",
"fields": {
"Description": {
"value": ""
},
"Name": {
"value": "H20 Chemical Drawing"
}
}
}
},
{
"type": "entity",
"id": "samplesContainer:ca05dfb4-d4cd-4910-a486-3ebaaf7982d7",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/samplesContainer:ca05dfb4-d4cd-4910-a486-3ebaaf7982d7"
},
"attributes": {
"type": "samplesContainer",
"eid": "samplesContainer:ca05dfb4-d4cd-4910-a486-3ebaaf7982d7",
"name": "Samples from Experiment",
"digest": "16509197",
"fields": {
"Description": {
"value": ""
},
"Name": {
"value": "Samples from Experiment"
}
}
}
},
{
"type": "entity",
"id": "text:df47fec8-d7fa-49a4-b2cf-c1468f1ad4e0",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:df47fec8-d7fa-49a4-b2cf-c1468f1ad4e0"
},
"attributes": {
"type": "text",
"eid": "text:df47fec8-d7fa-49a4-b2cf-c1468f1ad4e0",
"name": "Experiment Setup",
"digest": "29956862",
"fields": {
"Description": {
"value": ""
},
"Name": {
"value": "Experiment Setup"
}
}
}
},
{
"type": "entity",
"id": "text:18849188-9d5a-4064-a287-e4fe402028f8",
"links": {
"self": "https://snb.example.com/api/rest/v1.0/entities/text:18849188-9d5a-4064-a287-e4fe402028f8"
},
"attributes": {
"type": "text",
"eid": "text:18849188-9d5a-4064-a287-e4fe402028f8",
"name": "Experiment Conclusions",
"digest": "33833806",
"fields": {
"Description": {
"value": ""
},
"Name": {
"value": "Experiment Conclusions"
}
}
}
}
]
}
Response Breakdown:
Our response has three top level objects:
- “links”: The url of our Primary Resource which is represented by the
dataobject. This url will bring you to this same response if logged in to an account with the correct permissions. - “data”: This contains the data of our requested resource. In our case this is our requested experiment.
- "included:" The included array contains data related to our primary resource. In our example this is the user who created/edited/owns the Experiment, the user who edited the experiment, and the experiments children.
Breaking down the data object further:
“type”: This is the type of the data based on the API command used, in our case we asked for an Entity so our type is entity
“id”: The ID of our Primary Resource which is represented by the data object.
“links”: This contains an object, self, which holds the URL of our Primary Resource.
“attributes”: Attributes contains information about our Primary Resource.
“relationships”: This contains links and basic meta data, such as entity ID and type, of related resources. Related resources for our experiment include user data, the experiments children, and a convenient link to generate a PDF copy of the experiment.
Error Handling
Signals Notebook uses standard REST HTTP response codes to indicate success or failure of an API request. Typically a 2xx indicates success, a 4xx indicates an error on the client side, and 5xx indicates an error on Signal's side.
Typically errors are returned in a JSON dictionary containing four string values: a status code, a string code, a title, and a detailed description of the error.
{
"errors": [
{
"status": "",
"code": "",
"title": "",
"detail": ""
}
]
}Here is an example of a 401 Unauthorized error:
{
"status": "401",
"code": "Unauthorized",
"title": "User is unauthorized",
"detail": "No x-api-key is found in request header or the value of x-api-key is invalid."
}Concurrent Editing and use of "Digest" values
You will see in our example response, and most responses for entities, a digest value is included in the attributes object.
In the context of Signals API a "digest" refers to a mechanism used to ensure data integrity and manage concurrent updates. The "digest" is a unique identifier for a piece of data generated at the time of creation and updated when any change to the data occurs. This value is used to verify whether the data has been modified by another user during concurrent editing. This process is done automatically via the user interface and changes are reflected live for any end-user viewing the edited data.
To ensure integrity of the data when making use of the API for editing or creating data the following is an overview of how it is handled:
Initial State: When any entity is created/or successfully modified the server generates a digest for that entity.
User uses the API to create or edit data: A User (User A) attempting to make changes to the entity via API must also include the entities digest as part of their URL parameters along with the new or modified data.
Server Verification: Upon receiving the modified data and the original digest, the server recalculates the digest for the current state of the data. If the recalculated digest matches the original digest sent by User A, the server accepts the changes because it means that User A worked with the most recent version of the data.
Concurrent Modification: If another user (User B) had modified the same data in the meantime, the digest calculated by the server for the current state of the data will not match the original digest sent by User A. This mismatch indicates that there has been a concurrent modification.
Handling Conflicts: When a conflict is detected during interactions via the API, the server will return a 428 error code indicating a digest mismatch as seen below.
{
"errors": [
{
"status": "428",
"code": "DigestNotMatch",
"detail": "Digest mismatch."
}
]
}Forced override: API users can include force=true as part of their URL parameters to attempt to have force their changes regardless of it's current state. If unable to make the changes the API response will give the best error message it can for what failed. For example attempting for edit an entity that has been trashed with the force=true included in the URL parameters will result in the error below.
{
"errors": [
{
"status": "403",
"code": "PreconditionFailed",
"title": "Access forbidden.",
"detail": "This entity has been trashed or has trashed ancestor, it is read only now."
}
]
}Note: Always be mindful of using force=true as you may overwrite changes to entities that were recently edited.
By using digests in this way, the system can detect and manage concurrent edits to the same piece of data, helping to prevent conflicts and ensure data consistency.
Search
The Signals POST /entities/search endpoint is a powerful tool for getting the information you need out of Signals. Search requests require a JSON body as part of the API Call which represents the query you are searching for.
A very basic search for any Experiment named "My Experiment Name" in order of most recently change query would look as follows:
{
"query": {
"$and": [
{
"$match": {
"field": "type",
"value": "experiment"
}
},
{
"$match": {
"field": "name",
"value": "My Experiment Name"
}
},
{
"$match": {
"field": "isTemplate",
"value": false
}
}
]
},
"options": {
"sort": {
"modifiedAt": "desc"
}
}
}
Here we ensure we are looking at only entities whose type is experiment using the first $match in our query. We then ensure that we are not looking at templates by making sure the isTemplate field is false. Lastly with our options we set our sort to be based on the modifiedAt attribute in descending (desc) order.
Searchable Fields, Query Operators, and Search Options
Additional documentation on searchable fields, supported query operations, and search options can be accessed by system administrators in the System Configuration Guide.
Example Queries
Find all Experiments with a specific meta data field
In this example we are looking for all our Experiments that contain the custom field "Project Identifier" with a value of "Biology-100" sorted by their creation date.
{
"query": {
"$and": [
{
"$match": {
"field": "type",
"value": "experiment",
"mode": "keyword"
}
},
{
"$match": {
"field": "isTemplate",
"value": false
}
},
{
"$match": {
"field": "fields.Project Identifier",
"in": "tags",
"as": "text",
"value": "Biology-100",
"mode": "keyword"
}
}
]
},
"options": {
"sort": {
"createdAt": "desc"
}
}
}
Find all experiments with Benzene in their Chemical Drawing
In this example we are looking for all our Experiments that contain a Benzene molecule in their chemical drawing.
{
"query": {
"$and": [
{
"$match": {
"field": "type",
"value": "experiment",
"mode": "keyword"
}
},
{
"$match": {
"field": "isTemplate",
"value": false
}
},
{
"$child": {
"$chemsearch": {
"molecule": "C1=CC=CC=C1",
"mime": "chemical/x-daylight-smiles",
"options": "full=true"
}
}
}
]
}
}